
NIDRA v0.2.3 - super simple sleep scoring
https://github.com/paulzerr/nidraAn easy way to use powerful machine learning models to autoscore sleep recordings with excellent accuracy. No programming required, but Python endpoints are available. NIDRA can accurately score recordings from 2-channel EEG wearables such as ZMax (using ez6 and ez6moe models), as well as full PSG recordings (using U-Sleep 2.0 via sleepyland).
Download >> NIDRA for Windows 10/11 <<
Installation
Option 1: Standalone installer
The easiest way to use NIDRA on Windows is to download the portable self-extracting archive which requires no installation.
Note: due to the restrictive environment of Windows, you may see a "Search on app store" popup window. Click "No". You may also see the blue Smartscreen warning. In that case click "More info" and then "run anyway". Should this fail, try installing via pip (see below).
Option 2: Install with pip
It is highly recommended to create a clean virtual environment to install NIDRA. This prevents conflicts with other packages. Python 3.10 recommended.
Windows:
python -m venv nidra-env
nidra-env\Scripts\activate
pip install nidraMac/Linux:
python -m venv nidra-env
source nidra-env/bin/activate
pip install nidraOr install using Conda (Windows/Mac/Linux):
First install e.g., Miniconda.conda create -n nidra-env
conda activate nidra-env
pip install nidraLaunch the graphical interface:
nidraNote: If you installed via pip, the first time you run NIDRA, the necessary model files will be automatically downloaded from https://huggingface.co/pzerr/NIDRA_models/ (~152MB).
Option 3: Install from source
git clone https://github.com/paulzerr/nidra.git
cd NIDRA
pip install .Graphical user interface (GUI)
The GUI provides an intuitive, point-and-click option to score sleep recordings. The easiest way to launch the GUI is by downloading and starting the self-extracting archive (see above). Or, if you installed NIDRA as a Python package, you can launch it by opening your terminal or cmd and running the command: nidra
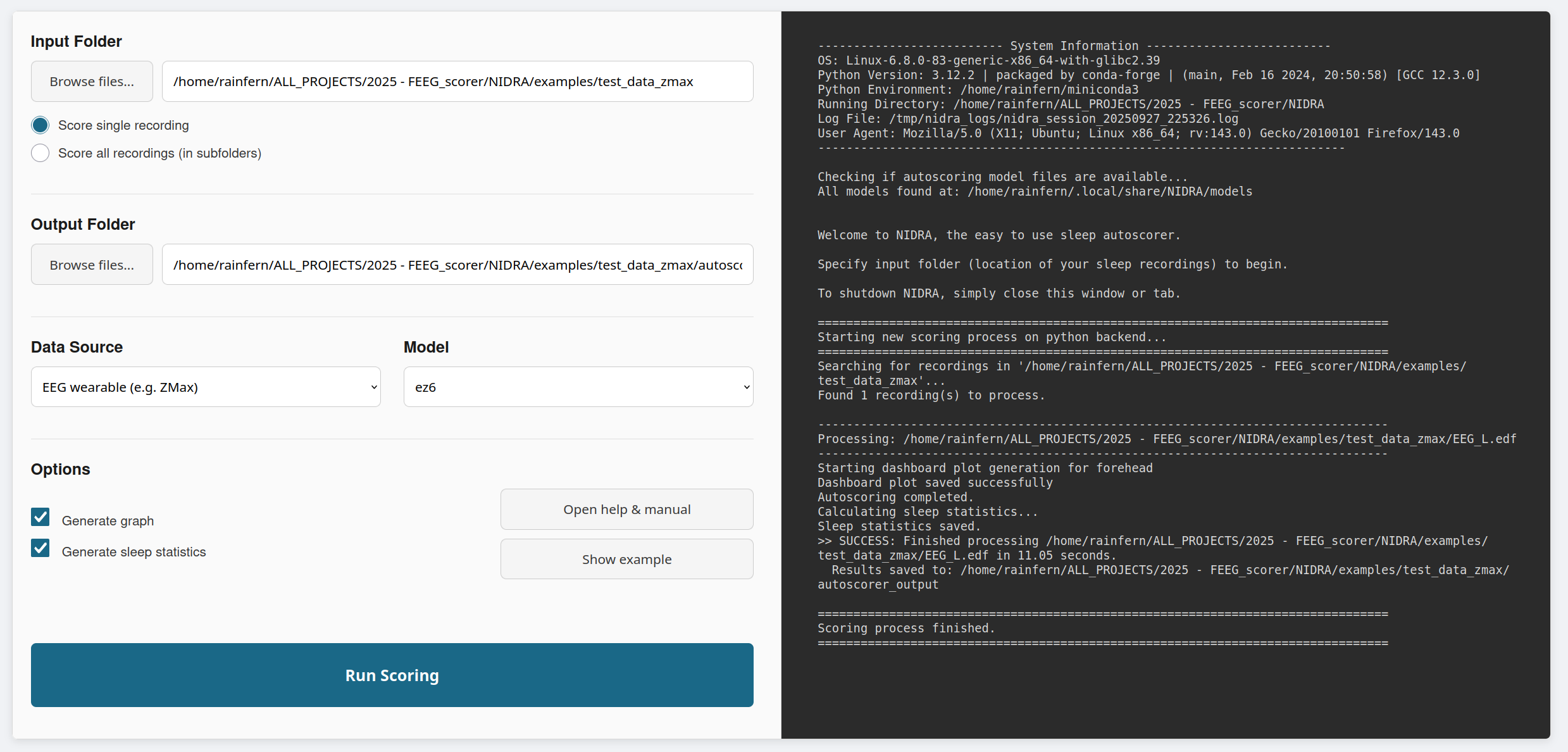
Fig.1 - Screenshot of the GUI.
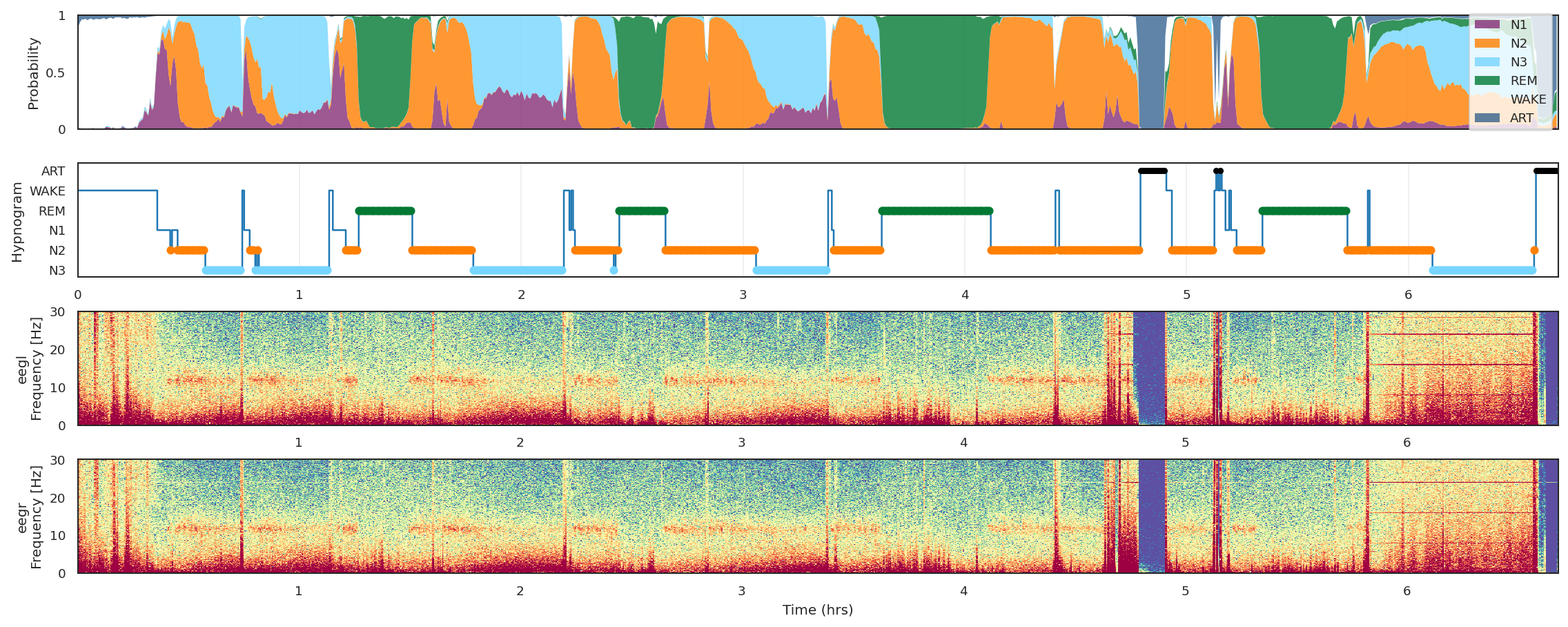
Fig.2 - Example output figure.
GUI Walkthrough
Step 1: Start the NIDRA GUI
For installation see top of this page. If installed via the standalone version, go to the directory where you extracted the files and double-click NIDRA.exe.
Note: due to the restrictive environment of Windows, you may see a "Search on app store" popup window. Click "No". You may also see the blue Smartscreen warning. In that case click "More info" and then "run anyway". Should this fail, try installing via pip (see installation section).
Alternatively, if you installed NIDRA as a Python package, open your terminal (On Windows: press CTRL+R, type in cmd, press Enter) and type nidra, then press Enter. The main application window will appear.
Step 2: Set input and output paths
Tell NIDRA where your data is located and where the results should go.
- Input: Click "Browse...". You can select a single recording file (EDF), a folder containing multiple recordings, or a text file (.txt) listing file paths. For processing multiple recordings, select the main project folder containing your recordings. NIDRA will automatically detect .edf files in subfolders. Please see "Preparing your data" section above for details.
- Output Directory: Specify the folder where NIDRA will save the scoring results. If you leave this blank, NIDRA will automatically create a new folder named
autoscorer_outputinside your input directory.
Step 3: Select recording type and model
- Data Source: Select the type of recording you have. Choose "EEG wearable" for 2-channel wearable data (e.g., ZMax), or "PSG" for standard polysomnography data.
- Model: The available models will change based on your recording type. For wearabnle EEG,
ez6moeis recommended for its high accuracy. This model will take approximately 30 seconds to score one night. Alternatively, useez6, which runs in about 5 seconds, but has slightly lower accuracy. For PSG,u-sleep-nsrr-2024(USleep 2.0) is the standard choice. This model uses all available EEG and EOG channels and will run one scoring process for each possible combination of single EEG and EOG channels, and then take a majority vote from each scoring process. If there are no EOG channels available, NIDRA will automatically use the EEG-onlyu-sleep-nsrr-2024_eegmodel.
Step 4: Run the analysis
Sleep stage output is always generated. You can choose whether to additionally generate classifier probability output, graphs, sleep statistics using the checkboxes. Click the "Run Autoscoring" button to begin. You can monitor the real-time progress in the console panel on the right. This log will show which files are being processed and report any warnings or errors.
Preparing your data
- NIDRA automatically detects your input format. When specifying a single .edf file, NIDRA will score that file. When specifying a folder, NIDRA will score all recordings in that folder and all of its subfolders. You can also input a text file that contains a list of file paths.
- When scoring PSG data, and if you use standard channel labels (e.g., 'EEG Fpz-Cz', 'EOG left', 'Fp1', 'O2-M1'), NIDRA uses these to identify channel types. If no clear channel names are provided, all channels are used and assumed to be EEG.
- When scoring ZMax or other EEG wearable data, your recordings may contain both channels in the same file, or you can have one channel per file. In the latter case, the file pairs need to be in the same folder and named *L.edf and *R.edf.
Structure for Forehead EEG (e.g., ZMax):
For ZMax EEG data in the original format, the left and right channel files must be in the same directory. These are typically named
EEG_L.edfandEEG_R.edf. However, you can also supply ZMax recordings (or data from any forehead EEG device) with both EEG channels in the same .edf file. NIDRA will automatically detect this.
forehead_study/ ├── subject_01/ | ├── EEG_L.edf | └── EEG_R.edf ├── subject_02/ | ├── night01_L.edf | └── night01_R.edf
Selecting channels
- You can optionally specify which channels to use for scoring. NIDRA will automatically detect EEG and EOG channels based on their names, and ignore other channels as they are not used by any of the models.
Model validation and performance
The models included in NIDRA have been rigorously validated against manually scored data from human experts. Below is a summary of their performance.
ezscore-f (ez6 and ez6moe) for wearable forehead EEG
ez6moe is recommended for its high accuracy. This model will take approximately 30 seconds to score one night. Alternatively, use ez6, which runs in about 5 seconds, but has slightly lower accuracy. This is mostly useful for testing purposes or real-time applications.
The confusion matrix below shows the ez6moe model's predictions (y-axis) versus the expert labels (x-axis). The diagonal represents correct classifications. The model shows high agreement with the expert scorer, particularly for Wake, N2, and REM sleep. For further details, please see the ezscore-f paper.
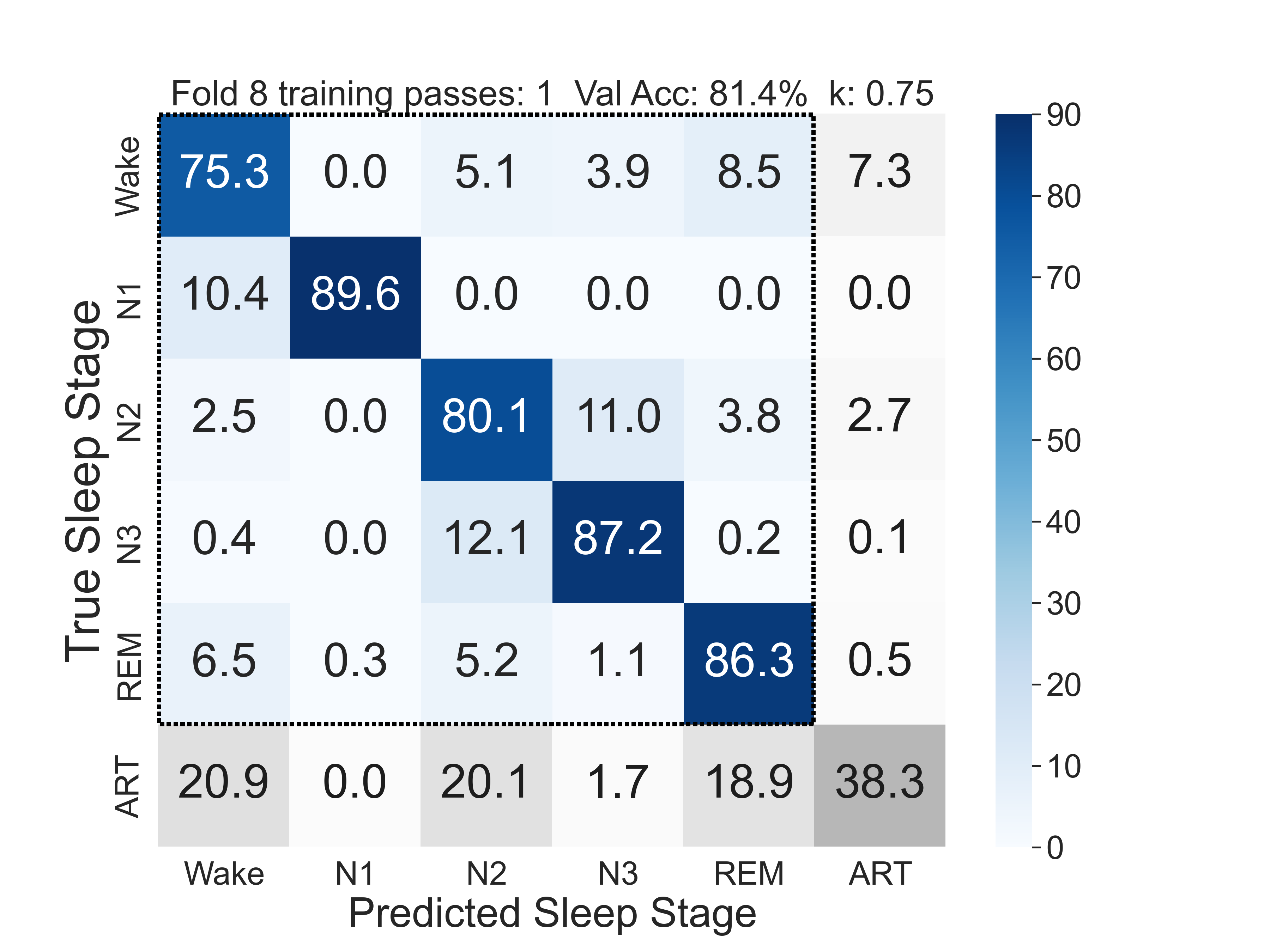
Fig.2 - Confusion matrix (vs. manually scored PSG) of the artefact-aware ez6moe model.
Key performance metrics, such as accuracy and Cohen's Kappa (a measure of inter-rater agreement), are comparable to the agreement levels seen between different human experts. For full details, please refer to the original publication.
U-Sleep for PSG
The U-Sleep model is a well-established, state-of-the-art algorithm for PSG sleep scoring. The version used in NIDRA (u-sleep-nsrr-2024) is a robust implementation trained on a large dataset. It demonstrates high performance across diverse populations and recording conditions. For detailed performance metrics, please see the original U-Sleep and SLEEPYLAND publications.
Understanding your results
| Output file | Description |
|---|---|
..._hypnogram.csv | A CSV with one integer per 30 s epoch (sleep stage code). |
..._hypnodensity.csv | Classifier probabilities per epoch; one column per stage (Wake, N1, N2, N3, REM, Artifact). |
..._figure.png | Plot with hypnogram, time-frequency spectrogram, and hypnodensity. |
..._sleep_statistics.csv | Summary metrics (TST, efficiency, time in each stage, etc.). |
| Sleep stage | Wake | N1 | N2 | N3 | REM | Artifact |
|---|---|---|---|---|---|---|
| Code | 0 | 1 | 2 | 3 | 5 | 6 |
Python endpoints
Example 1: Scoring a single PSG recording (minimal example)
import NIDRA
scorer = NIDRA.scorer(
type = 'psg',
input = '/path/to/recording.edf'
)
scorer.score()
Example 2: Scoring a single wearable-EEG recording File (specifying optional parameters)
import NIDRA
scorer = NIDRA.scorer(
type = 'forehead',
input = '/path/to/recording/',
output = '/path/to/output/folder/',
model = 'ez6moe',
channels = ['eegl','eegr'],
hypnogram = True,
hypnodensity = True,
plot = True,
)
hypnogram, probabilities = scorer.score()
Example 3: Scoring multiple recordings
You can score multiple recordings in one go by simply specifying a folder path, which contains recordings or subfolders with recordings. NIDRA will scan subfolders recursively and score all edf files present.
You can also specify the path of a .txt file that contains one path to a recording (or folder containing one or more recordings) per line.
import NIDRA
scorer = NIDRA.scorer(
type = 'psg',
input = '/path/to/folder/'
)
scorer.score()
Example 4: Scoring in-memory data (e.g. real-time application)
You can also score data that you already have in memory as an array. This is useful for real-time applications or custom data loading pipelines. When scoring PSG data from an array, you must provide the sampling frequency (sfreq). Providing channel names is recommended but optional; if not provided, they will be auto-generated. All channels are then assumed to contain EEG data. By default, no output files are generated, but this can be enabled if desired.
# create some dummy PSG data
import numpy as np
sfreq = 256
channels = ['F3-A2', 'C4-A1', 'O2-A1', 'EOG-L']
n_samples = sfreq * 60 * 60 # 1 hour of data
dummy_data = np.random.randn(len(channels), n_samples)
import NIDRA
scorer = NIDRA.scorer(
type='psg',
input=dummy_data,
sfreq=sfreq,
channels=channels
)
hypnogram, probabilities = scorer.score()
Reference
Single user-facing entry point NIDRA.scorer(...). The data source (PSG or wearable EEG) is selected by scorer_type. Then call .score() to run inference. This unified reference clarifies which parameters are required, optional, and their defaults across PSG/Forehead and file/array scenarios.
Entrypoint
NIDRA.scorer(scorer_type: str, **kwargs) -> Scorer
Scorer.score(plot: bool = False) -> tuple[numpy.ndarray, numpy.ndarray]Returns
hypnogram— 1D numpy array of integer stage codes for each 30 s epoch (see Sleep Stage Key)probabilities— 2D array [n_epochs, n_classes] with per-epoch, per-class probabilities
Output files (when enabled)
..._hypnogram.csv— sleep stage code per epoch..._hypnodensity.csv— per-epoch class probabilities..._figure.png— plot (ifplot=Trueand file writing enabled)
| Parameter | Type | Default | Required? | Description |
|---|---|---|---|---|
type |
str |
— | Yes | Type of data source: 'psg' (full PSG with EEG and optionally EOG data) or 'forehead' (wearable 2-channel EEG). |
input |
str | Path | numpy.ndarray |
— | Yes |
Location of recording(s) to score. File path (str/Path): Path to recording file (EDF/BDF), folder, or .txt file with one path per line. Array (numpy.ndarray): In-memory signal data. PSG array: shape (C, N) — any number of channels C. Forehead array: shape (2, N) — exactly 2 channels required. |
output |
str |
input/autoscorer_output |
— | Directory for outputs. Defaults to autoscorer_output in input folder. Required if using in-memory data and saving files. |
sfreq |
float |
— | Required when input is an array |
Sampling frequency (Hz) for array input. |
channels |
list[str] |
— | — | Define list of channels to use for scoring. In case of "forehead" data source, provide exactly two channels, unless scoring original ZMax file pairs, in which case do not provide any channels. |
model |
str |
PSG: 'u-sleep-nsrr-2024'Forehead: 'ez6'
|
— |
Selects the model to use. PSG: Default is 'u-sleep-nsrr-2024' (USleep 2.0). Automatically falls back to EEG-only version (..._eeg) if no EOG channels are detected.Forehead: Use 'ez6' (fast) or 'ez6moe' (higher accuracy).
|
hypnogram |
bool | None |
True when input is a file;False when input is an array
|
— | Controls whether hypnogram CSV is written to output. |
hypnodensity |
bool |
False |
— | Controls whether probability CSV is written to output. |
plot |
bool |
False |
— | Controls whether summary plot is written to output. |
Additional semantics
inputcan handle file paths, folders or in-memory arrays.- Forehead array input must be exactly two channels (2×N).
- Forehead file input: pass the LEFT or RIGHT channel EDF (other inferred) OR a single EDF containing both channels.
- Forehead single-file: optionally specify
channels; otherwise the first two channels are used. - All models operate on 30-second epochs; outputs align to full epochs.
Machine-readable schema
{
"entrypoint": "NIDRA.scorer",
"parameters": {
"type": { "type": "string", "enum": ["psg", "forehead"], "required": true },
"output": { "type": "string", "required": false, "default_logic": "autoscorer_output folder in input directory" },
"input": {
"type": "string | array[number]",
"required": true,
"description": "Path to file/folder/.txt list OR numpy array (channels x samples)"
},
"sfreq": { "type": "number", "required_if": "input is array" },
"channels": { "type": "array[string]", "required": false },
"model": {
"type": "string",
"required": false,
"defaults": { "psg": "u-sleep-nsrr-2024", "forehead": "ez6" }
},
"hypnogram": {
"type": "boolean|null",
"required": false,
"default_logic": "True if input is file, False if array"
},
"hypnodensity": { "type": "boolean", "required": false, "default": false },
"plot": { "type": "boolean", "required": false, "default": false }
},
"returns": {
"hypnogram": { "type": "array[int]", "epoch_sec": 30 },
"probabilities": { "type": "array[array[number]]", "shape": "[n_epochs, n_classes]" }
},
"artifacts": [
"hypnogram.csv", "probabilities.csv", "figure.png (when plot=True)"
]
}FAQ & troubleshooting
Installation issues
Q: I see a warning when trying to run the Windows .exe (Smartscreen or "Search on app store").
A: Due to the restrictive environment of Windows, you may see a "Search on app store" popup window. Click "No". You may also see the blue Smartscreen warning. In that case click "More info" and then "run anyway". Should this fail, try installing via pip (see Installation section).
Q: I'm having trouble installing with pip. What can I do?
A: Here are a few common troubleshooting steps:
- 1. Update packages: Ensure you have the latest versions of pip, setuptools, and wheel, as outdated versions can cause issues. You can update them by running:
pip install --upgrade pip setuptools wheel - 2. Use a clean virtual environment: Conflicts with other installed packages are a common source of errors. Creating a fresh virtual environment ensures that NIDRA's dependencies are installed in an isolated space.
- 3. Try installing with conda: Conda's package management is often better at resolving complex dependencies. Follow the Conda installation instructions in the Installation section.
GUI issues
Q: The GUI window is not appearing when I run nidra.
A: Check the terminal for any error messages. This could be due to a missing dependency. Try reinstalling NIDRA in a clean virtual environment.
Q: Why is the "Run Autoscoring" button disabled?
A: The button is disabled until you have selected a valid recording to sco.
Scoring errors
Q: I got an error about "missing channels" or "could not find required channels".
A: This is a common error that occurs when the channel labels in your EDF file do not match what the model expects, or when you've selected the wrong "Data Source". For example, selecting "PSG" for a forehead EEG file will cause this error. Double-check your data source selection and ensure your EDF channel labels are standard.
General questions
Q: Can I trust the results?
A: The models in NIDRA are validated and perform at a level comparable to human experts. However, like any automated algorithm, they are not perfect. It is always good practice to visually inspect the generated hypnogram plot for any obvious anomalies, especially for noisy or unusual recordings.
How to cite NIDRA
If you use NIDRA in your research, please cite both the NIDRA software itself and the paper for the specific model you used.
1. Citing the NIDRA Software
Please cite this repository to ensure reproducibility:
Zerr, P. (2025). NIDRA: super simple sleep scoring. GitHub. https://github.com/paulzerr/nidra
2. Citing the Scoring Model
If you used the ez6 or ez6moe models:
Coon WG, Zerr P, Milsap G, et al. (2025). ezscore-f: A Set of Freely Available, Validated Sleep Stage Classifiers for Forehead EEG. bioRxiv. doi: 10.1101/2025.06.02.657451.
If you used the u-sleep-nsrr-2024 model:
Please cite the original U-Sleep paper and the SLEEPYLAND paper for the re-trained model weights:
Perslev, M., et al. (2021). U-Sleep: resilient high-frequency sleep staging. NPJ digital medicine. Rossi, A. D., et al. (2025). SLEEPYLAND: trust begins with fair evaluation of automatic sleep staging models. arXiv preprint.
Attribution
ez6 and ez6moe models were developed by Coon et al., see:
Coon WG, Zerr P, Milsap G, Sikder N, Smith M, Dresler M, Reid M.
ezscore-f: A Set of Freely Available, Validated Sleep Stage Classifiers for Forehead EEG.
https://www.biorxiv.org/content/10.1101/2025.06.02.657451v1
github.com/coonwg1/ezscore
U-Sleep models were developed by Perslev et al., see:
Perslev, M., Darkner, S., Kempfner, L., Nikolic, M., Jennum, P. J., & Igel, C. (2021).
U-Sleep: resilient high-frequency sleep staging. NPJ digital medicine
https://www.nature.com/articles/s41746-021-00440-5
https://github.com/perslev/U-Time
The U-Sleep model weights used in this repo were re-trained by Rossi et al., see:
Rossi, A. D., Metaldi, M., Bechny, M., Filchenko, I., van der Meer, J., Schmidt, M. H., ... & Fiorillo, L. (2025).
SLEEPYLAND: trust begins with fair evaluation of automatic sleep staging models. arXiv preprint arXiv:2506.08574.
https://arxiv.org/abs/2506.08574v1
https://github.com/biomedical-signal-processing/sleepyland
License
This project is licensed under the MIT License. See the LICENSE file for details.
Contact
For questions, bug reports, or feedback, please contact Paul Zerr at zerr.paul@gmail.com or open a github issue.